IOCBIO Kinetics is a cross-platform application for analysis of different traces, as described by its plugins. While originally developed for analysis of enzyme kinetics, the other types of traces can be analyzed as well. It is designed to analyze traces where some measured parameter depends on one other parameter, such as time or space. For example, respiration rate measurements via following oxygen concentration in time and its changes induced through addition of metabolites.
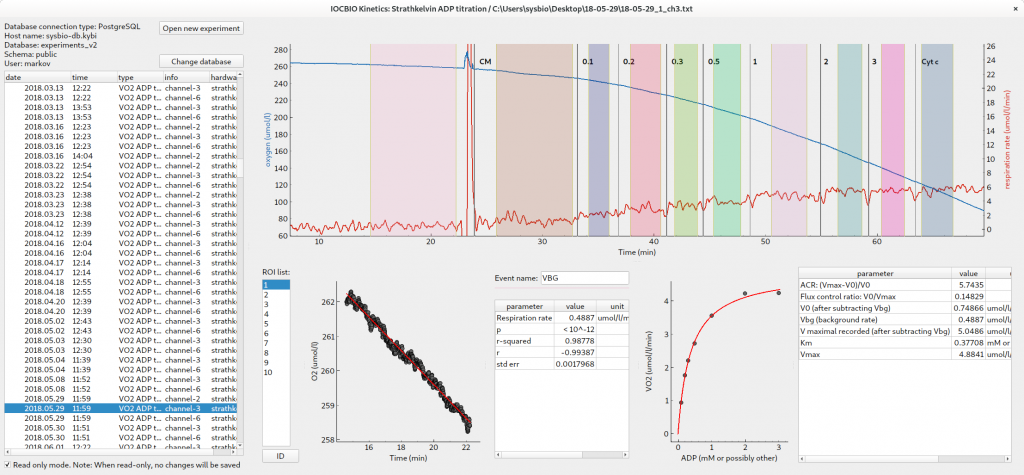
The analysis of experimental traces is built as a pipeline, with the data imported from the experiment file, regions of interest automatically generated or set by user, data fitted and analyzed. Communication between different plugins is done through the database backend with the analysis results stored in the database. Software is modular with the new modules added easily to fit the new types of experiments.
Links
- Homepage: https://iocbio.gitlab.io/kinetics
- Project page: https://gitlab.com/iocbio/kinetics
- Demos: https://www.youtube.com/channel/UCAyvqIEqVARCmtQ_5XQYJaQ
- Issues with the software and requests for help: https://gitlab.com/iocbio/kinetics/issues. Please use the public issues when you want to get assistance with installation, reading your data, suggest extension or improvement of the software. By doing it openly, you can help others with the same problems or suggestions.
Citation
Please cite paper describing software if you use it:
Vendelin, M., Laasmaa, M., Kalda, M., Branovets, J., Karro, N., Barsunova, K., & Birkedal, R. (2020). IOCBIO Kinetics: An open-source software solution for analysis of data traces. PLOS Computational Biology, 16(12), e1008475. https://doi.org/10.1371/journal.pcbi.1008475