Karo J, Peterson P, Vendelin M
J. Biol. Chem. 2012 Mar;287(10):7467-76
PMID: 22241474
Abstract
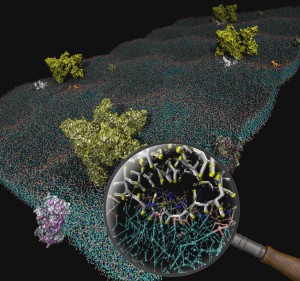
Interaction between mitochondrial creatine kinase (MtCK) and adenine nucleotide translocase (ANT) can play an important role in determining energy transfer pathways in the cell. Although the functional coupling between MtCK and ANT has been demonstrated, the precise mechanism of the coupling is not clear. To study the details of the coupling, we turned to molecular dynamics simulations. We introduce a new coarse-grained molecular dynamics model of a patch of the mitochondrial inner membrane containing a transmembrane ANT and an MtCK above the membrane. The membrane model consists of three major types of lipids (phosphatidylcholine, phosphatidylethanolamine, and cardiolipin) in a roughly 2:1:1 molar ratio. A thermodynamics-based coarse-grained force field, termed MARTINI, has been used together with the GROMACS molecular dynamics package for all simulated systems in this work. Several physical properties of the system are reproduced by the model and are in agreement with known data. This includes membrane thickness, dimension of the proteins, and diffusion constants. We have studied the binding of MtCK to the membrane and demonstrated the effect of cardiolipin on the stabilization of the binding. In addition, our simulations predict which part of the MtCK protein sequence interacts with the membrane. Taken together, the model has been verified by dynamical and structural data and can be used as the basis for further studies.