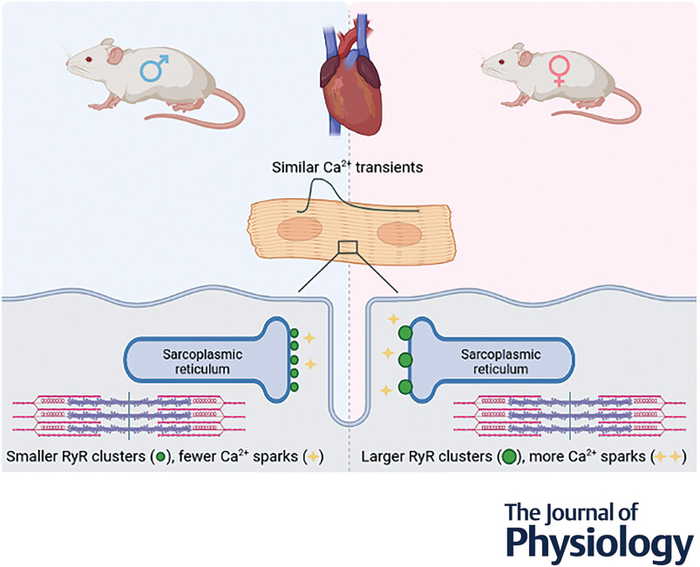
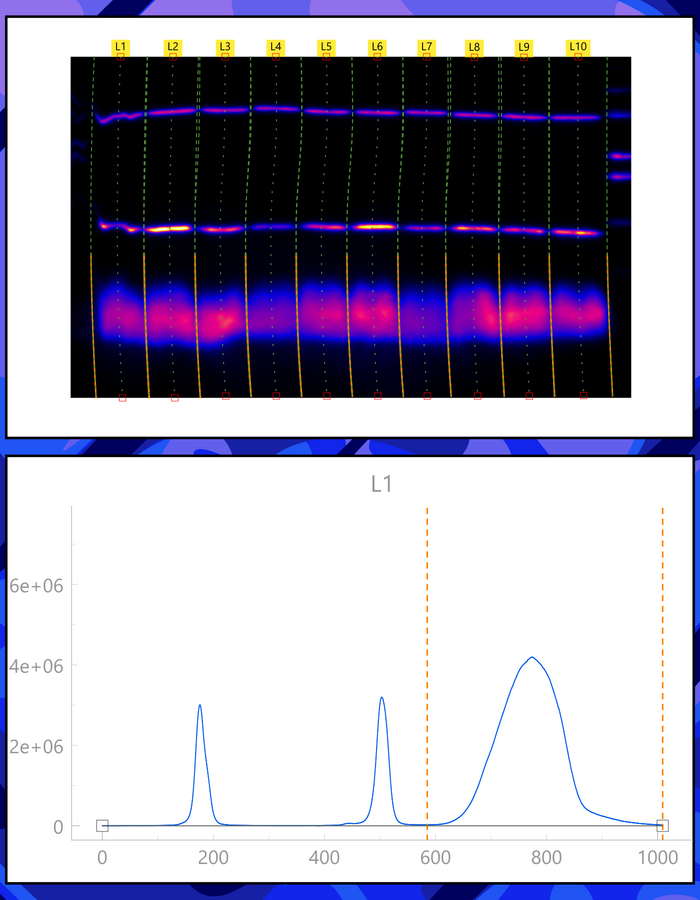
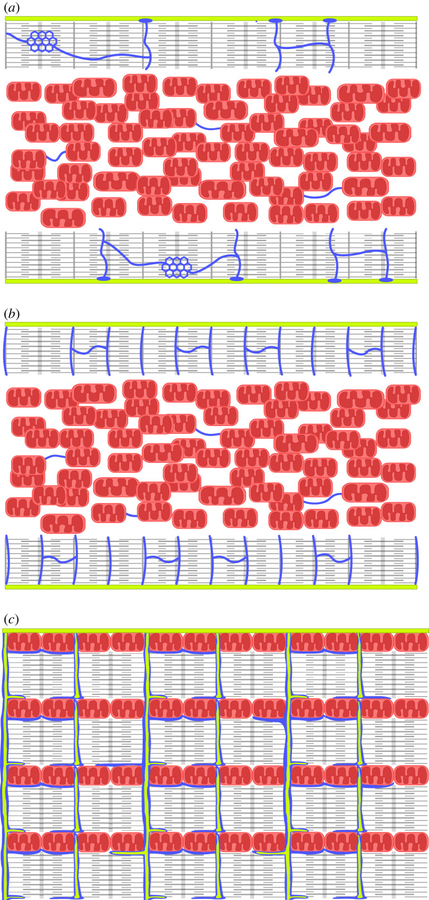
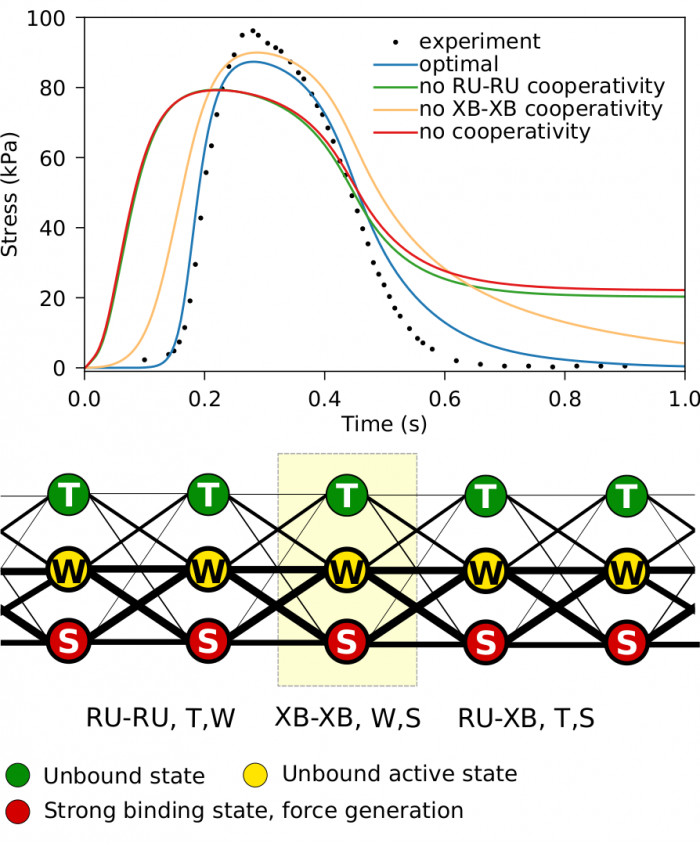
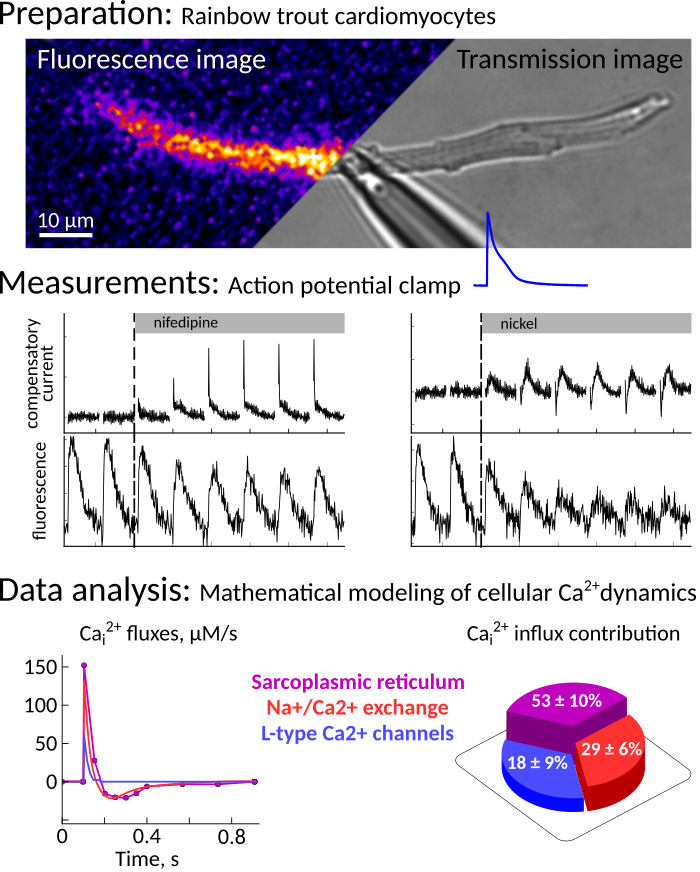
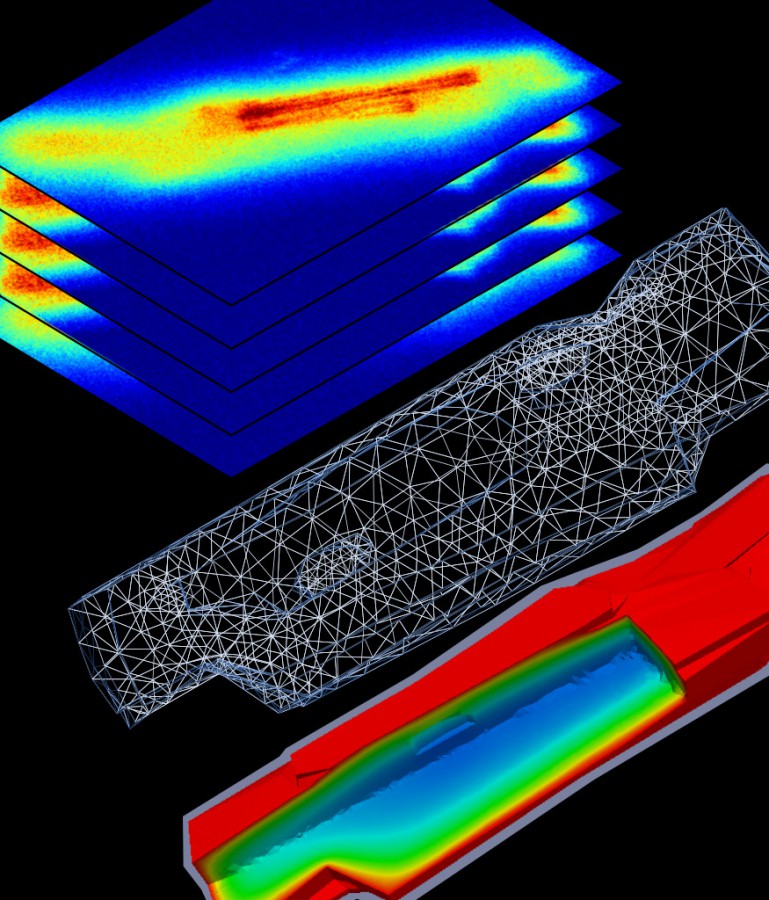
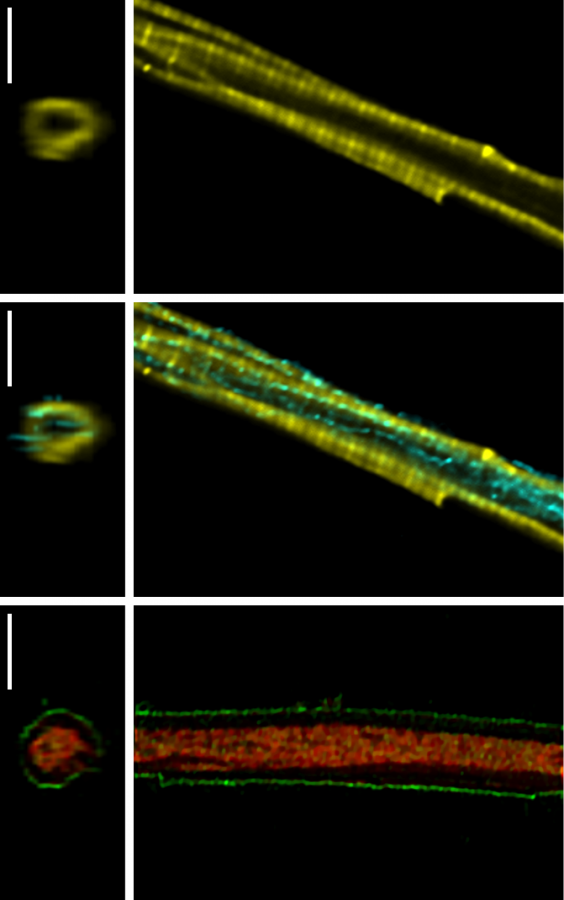
The Laboratory of Systems Biology is a part of the Department of Cybernetics, School of Science, Tallinn University of Technology.
AIM
The main aim of the laboratory is to study regulation of intracellular processes and understand functional influences of intracellular interactions.
For this, a mixture of experimental and theoretical approaches are used.
SPECIFIC SCIENTIFIC INTERESTS, PROJECTS
- Intracellular compartmentation and diffusion restrictions, influence on bioenergetics.
- How the fine structure of mitochondrion is influencing the dynamics of oxidative phosphorylation?
- Regulation of energy transfer in different types of cells.
- Metabolic stability of heart muscle cells.
- Interaction between mechanics and energetics of the heart.
APPROACH
We use interdisciplinary approaches to tackle questions in cardiac physiology. For that, we have formed a team of researchers with backgrounds in biophysics, biology, and applied mathematics/physics. As a result, we are able to approach scientific questions on different scales, from organ to molecular level, using combinations of different experimental and theoretical techniques. When needed, we find new ways to characterize the data, develop new mathematical models, build new hardware and program it to carry out novel experimental protocols. For example, we have developed mitochondrial positioning analysis in 2D and 3D, and built a confocal microscope to study diffusion in the cell. Among the experimental techniques routinely used in the laboratory are
- kinetic measurements,
- fluorescence correlation spectroscopy based techniques,
- confocal, super-resolution, TIRF, and wide-field fluorescence imaging,
- controlled single cell force measurements,
- electrophysiology, patch clamping.
We have developed models
- to study energetics and mechanics of the left ventricle as well as intracellular diffusion using finite element method,
- solved partial differential equations to analyze cross-bridge mechanoenergetics,
- used molecular dynamics to analyze interaction between proteins and mitochondrial membrane,
- designed multiple other models and computational techniques to analyze the experimental data.
The results of our work are distributed through publications and the open-source software that we develop.
NETWORKS
The laboratory participates in the following research networks:
- EUropean network to tackle METAbolic alterations in HEART failure, [CA COST Action CA22169, 2023-2027],
- Realising the therapeutic potential of novel cardioprotective therapies, [CA COST Action CA16225, 2017-2022],
- Mitochondrial mapping: Evolution – Age – Gender – Lifestyle – Environment, [CA COST Action CA15203, 2016-2021],
SUPPORT
The research in laboratory is financed by
- Estonian Research Council [ETAg, 2021-2025, PRG1127]
- Estonian Research Council [ETAg, 2023-2027, PSG832]
Previous funding:
- Estonian Research Council [ETAg, 2015-2020]
- European Regional Development Fund [CENS, 2011-2015],
- Wellcome Trust [Aug 2007-Jan 2014],
- Estonian Ministry of Education and Research [2008-2013],
- MOBILITAS Postdoctoral Research Grants [2009-2015],
- Estonian Science Foundation [2008-2012].
STUDENTS
See list for proposed students topics under Positions.
CONTACT
Head: Marko Vendelin Laboratory of Systems Biology Department of Cybernetics School of Science Tallinn University of Technology Akadeemia 15 12618 Tallinn Estonia Phone: +372 620 4169 https://sysbio.ioc.ee